Tools
The approaches used to generate data for this project involve:
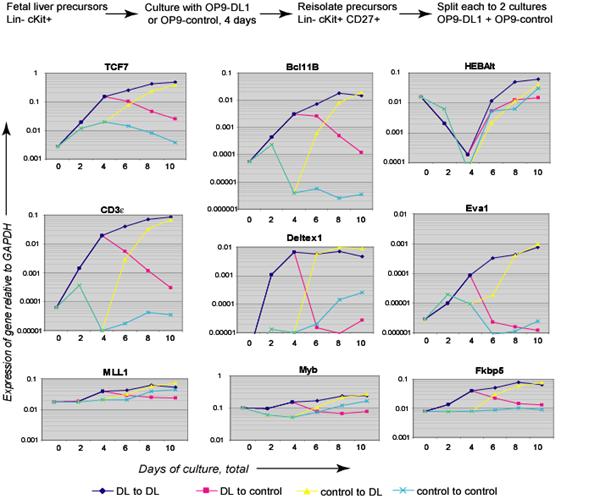
- Multigene correlation analysis across finely distinguished developmental stages in vivo
- Experimental manipulation of early T-cell development in vitro
- Perturbation analysis by gain and loss of transcription factor function at particular stages, correlated with effects on development and gene expression
- Perturbation analysis by addition or removal of Notch ligands from environment
- Genome-wide prediction of transcription factor-target linkages
- Chromatin immune precipitation and genome-wide analysis to assess active cis-regulatory sites
Sources of material:
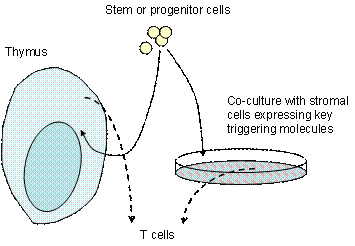 We use adult or
fetal mouse hematopoietic stem cells or defined immature cell types
from the thymus and grow them in culture dishes to control the
developmental environment of HSCs. These cells can develop
into T cells if and only if the environment expresses Notch ligands.
We use adult or
fetal mouse hematopoietic stem cells or defined immature cell types
from the thymus and grow them in culture dishes to control the
developmental environment of HSCs. These cells can develop
into T cells if and only if the environment expresses Notch ligands.
We use a variety of experimental technologies to study T cell development.
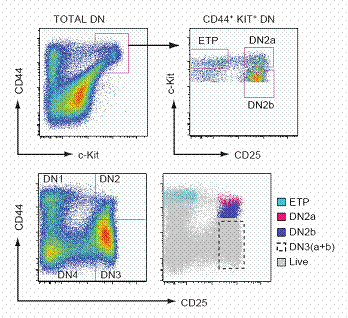
Using Fluorescence Assisted Cell Sorting (FACS), we can track the development of individual cells.
FACS analysis has led us to extremely fine-scale resolution of stages, subdividing the conventional DN1 – 4 stages for precise temporal correlations.