HOME | PEOPLE| RESEARCH| PUBLICATIONS| NEWS AND EVENTS| INTERNAL
Scientific Cores | Collaborators | Sponsors | Scientific Advisory Board
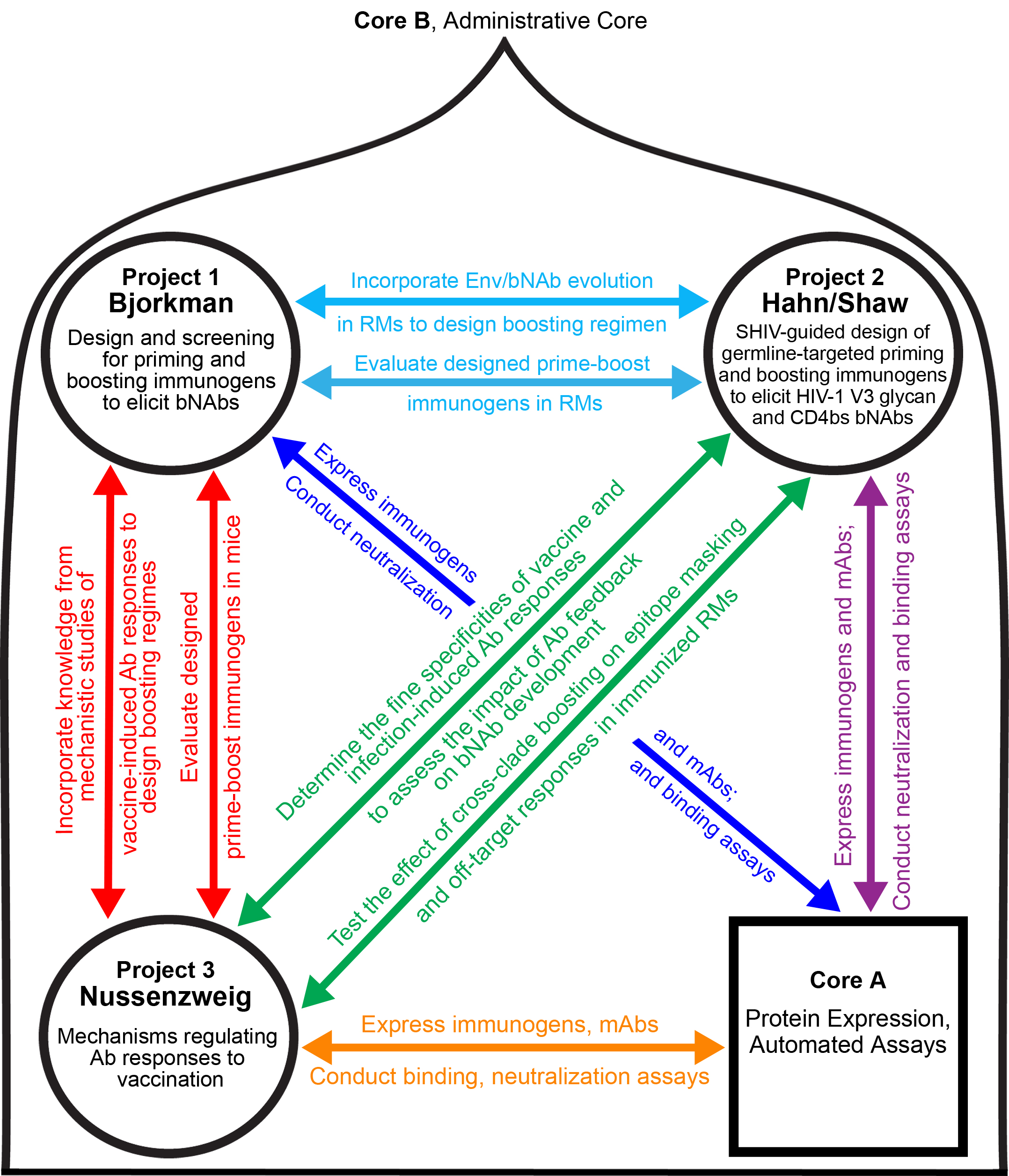
Projects:
Project 1. (Bjorkman) Design and screening for priming and boosting immunogens to elicit bNAbs. We engineered Env trimer immunogens as multimers on protein nanoparticles to induce bNAbs against three epitopes: (1) V3 (2) CD4bs, and (3) V1V2 (in our V3/CD4bs/V1V2 triple immunogen). These immunogens elicit heterologous neutralizing Ab responses in iGL transgenic mice and wt animals, providing a scaffold to further modify Envs to improve epitope targeting. Boosting regimens designed through understanding Env evolution during pressure from elicited Abs are required to mature primed Abs into potent bNAbs. We will collaborate with Drs. Hahn and Shaw to use Env sequence information from immunogen-primed RMs that are then infected with a lineage-maturing SHIV engineered to match or derive from a boosting immunogen such that the virus will “pick up” where the prime/boost regimen left off in terms of maturing the desired Ab response. We will also collaborate Dr. Weissman (Project 2) to design mRNA-LNP immunogens and with Dr. Nussenzweig (Project 3) to investigate whether boosting is impaired by high titer Abs elicited by GL-targetin
Project 2. (Hahn/Shaw) SHIV-guided design of priming and boosting immunogens to elicit HIV-1 V3 glycan and CD4bs bNAbs. SHIV-infected RMs develop HIV-1 bNAbs via Env-Ab coevolutionary pathways that recapitulate events in HIV-1 infected humans. Project 2 focuses lineage-based immunogen design for two of the most advanced vaccine targets: the V3 glycan patch and the CD4bs, both of which were developed as vaccine targets (the RC1 and IGT2 immunogens) in the currently funded HIVRAD. Specifically, Project 2 will generate GL-targeted SHIVs bearing Envs with selective glycan deletions and other mutations surrounding the V3 glycan and CD4bs epitopes so as to immunofocus early B cell responses to these bNAb epitopes. Project 2 will also develop lineage-maturing SHIVs designed to boost and affinity-mature bNAb responses that are primed by GL-targeted Env trimer nanoparticles or mRNA-LNP vaccines. In both instances, we will decipher SHIV Env-Ab co-evolution leading to neutralization breadth where, unlike in humans, we can analyze multiple animals infected by the same SHIVs and identify maturation pathways that are shared among different outbred animals. Using newly developed bioinformatic and phylogenetic tools, we will develop a “molecular blueprint” for the design of a lineage-based, SOSIP Env-SpyCatcher nanoparticle or mRNA-LNP expressed stabilized membrane Env immunization regimen that recapitula
Project 3. (Nussenzweig) Mechanisms regulating Ab responses to vaccination. To aid the design of better boosting immunogens by the Bjorkman and Hahn/Shaw labs, the Nussenzweig lab will conduct experiments in wt and transgenic mice and dissect mechanisms that lead to off-target epitope responses in sequential immunization. Preliminary data in mice and humans from the Nussenzweig lab indicates that Ab feedback and the quality and quantity of the T follicular helper cell response affect recruitment and affinity maturation of germinal center (GC) B cells. Experiments were performed with passively infused monoclonal Abs (mAbs) in immunologically intact mice and humans, illustrating that Ab feedback favors emergence of off-target responses by masking target epitopes and that memory B cells emerging from GCs are selected by a mechanism favoring Ab repertoire diversity over affinity. Using HIV-1 immunogens designed by the Bjorkman lab, the Nussenzweig lab will explore how polyclonal Ab responses alter the outcome of sequential immunization with the aim to improve on-target responses during boosting. The concepts that will be delineated from these experiments will then be tested in RM models by the Hahn/Shaw laboratories.